The burden of drug-resistant infections continues to threaten our arsenal of antibiotics. Despite the rise in genomic sequencing of clinical isolates, there is a lack of evidence-based guidelines and standardization when it comes to reporting results of genome sequencing. During May 2022, we surveyed clinicians, diagnosticians, and public health officials in Ontario (Canada) using LimeSurvey in order to assess the current state of genomic sequencing reporting, the current challenges, and recommendations for the content and design of genome reports. The survey was sent electronically and consisted of both qualitative and quantitative questions. The survey was divided into three sections:
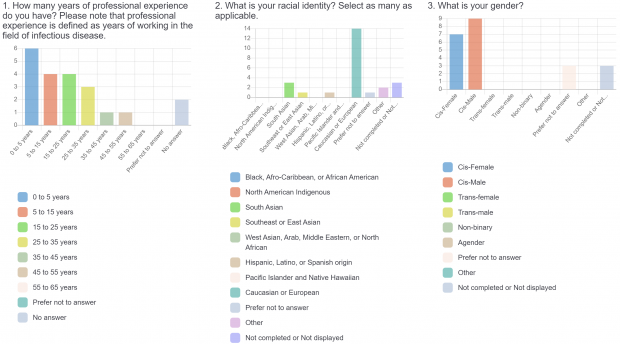
- Demographic information (years of professional experience, gender identity, racial identity, public health unit, professional role).
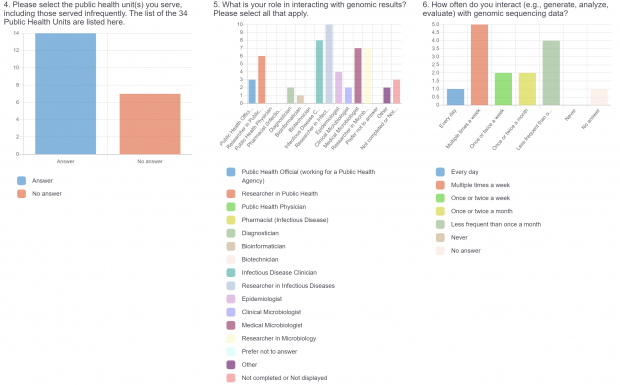
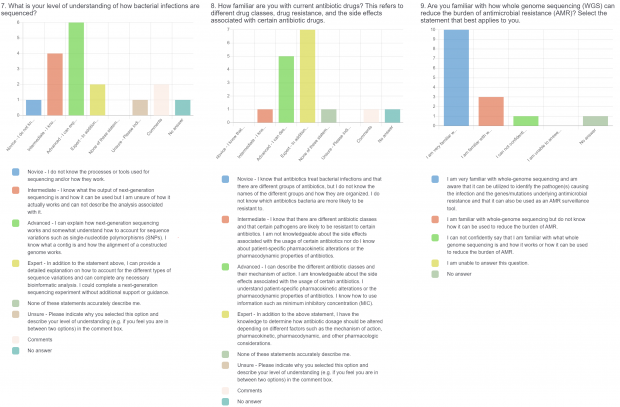
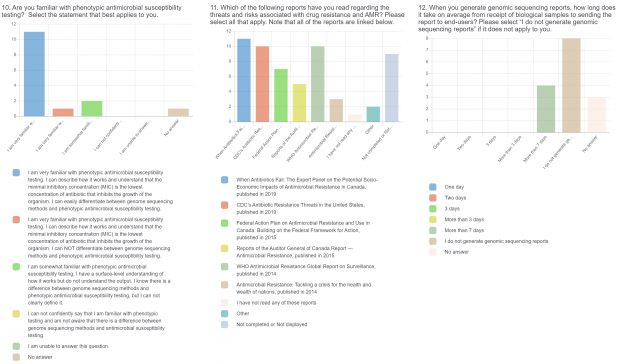
- Current practices associated with genomic sequencing reporting and knowledge about sequencing, antimicrobial resistance (AMR), and genomics.
- Improvements to current reporting practices.
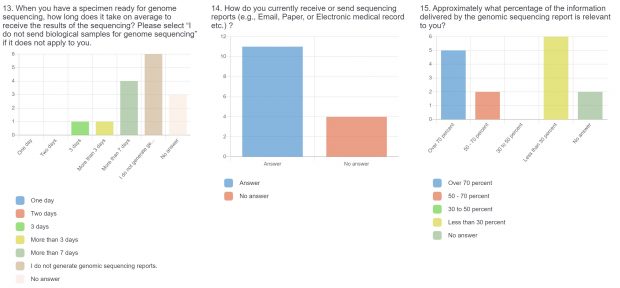
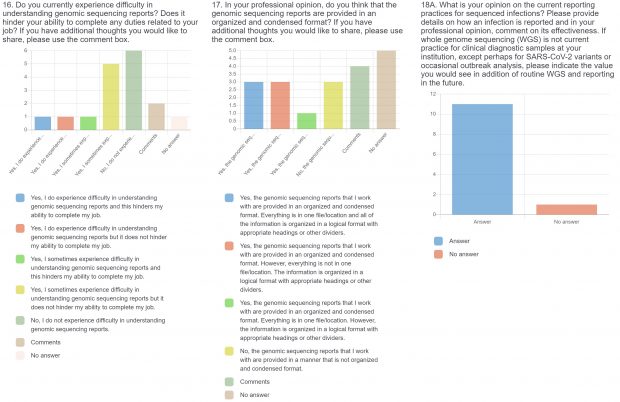
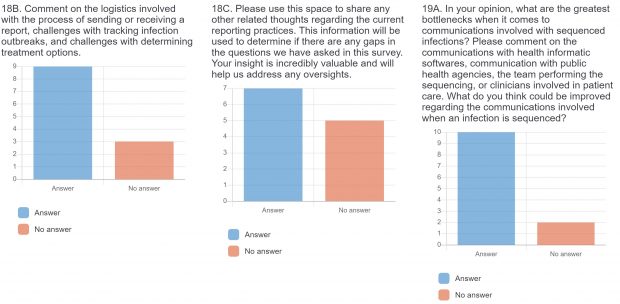
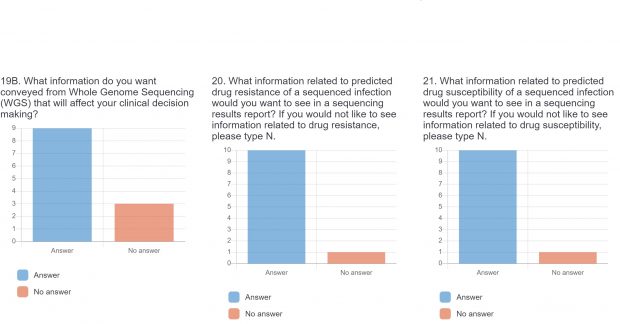
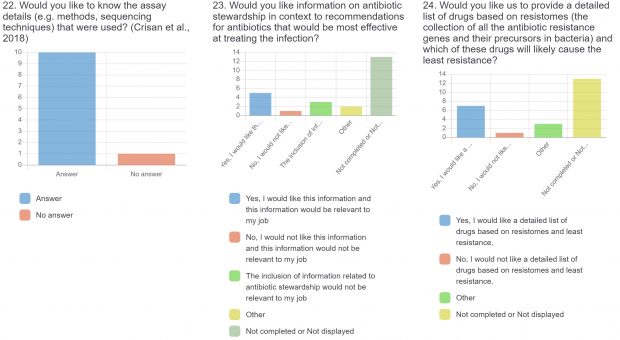
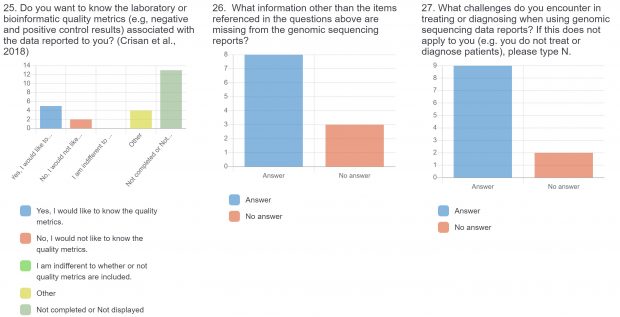
The raw results of the survey are displayed below. Statistical analysis of quantitative or categorical questions is ongoing. For qualitative questions, only response rates are shown – written responses will be analyzed separately. All findings will be included in a final publication.
Survey responses did not contain any identifying information about respondents. This research was approved by the Hamilton Integrated Research Ethics Board (HiREBProject#: 14152).
To cite these results: Dave, M. & A.G. McArthur. 2022. MACGENOM Raw Data – Understanding the current state of genomic reporting for drug resistant infections and creation of automated genomic sequencing reports. https://mcarthurbioinformatics.ca/macgenom/
You can alternatively download a PDF version of these results.
For more information, please contact Mugdha Dave of McMaster University at: macgenom@mcmaster.ca.
click on images for a magnified view