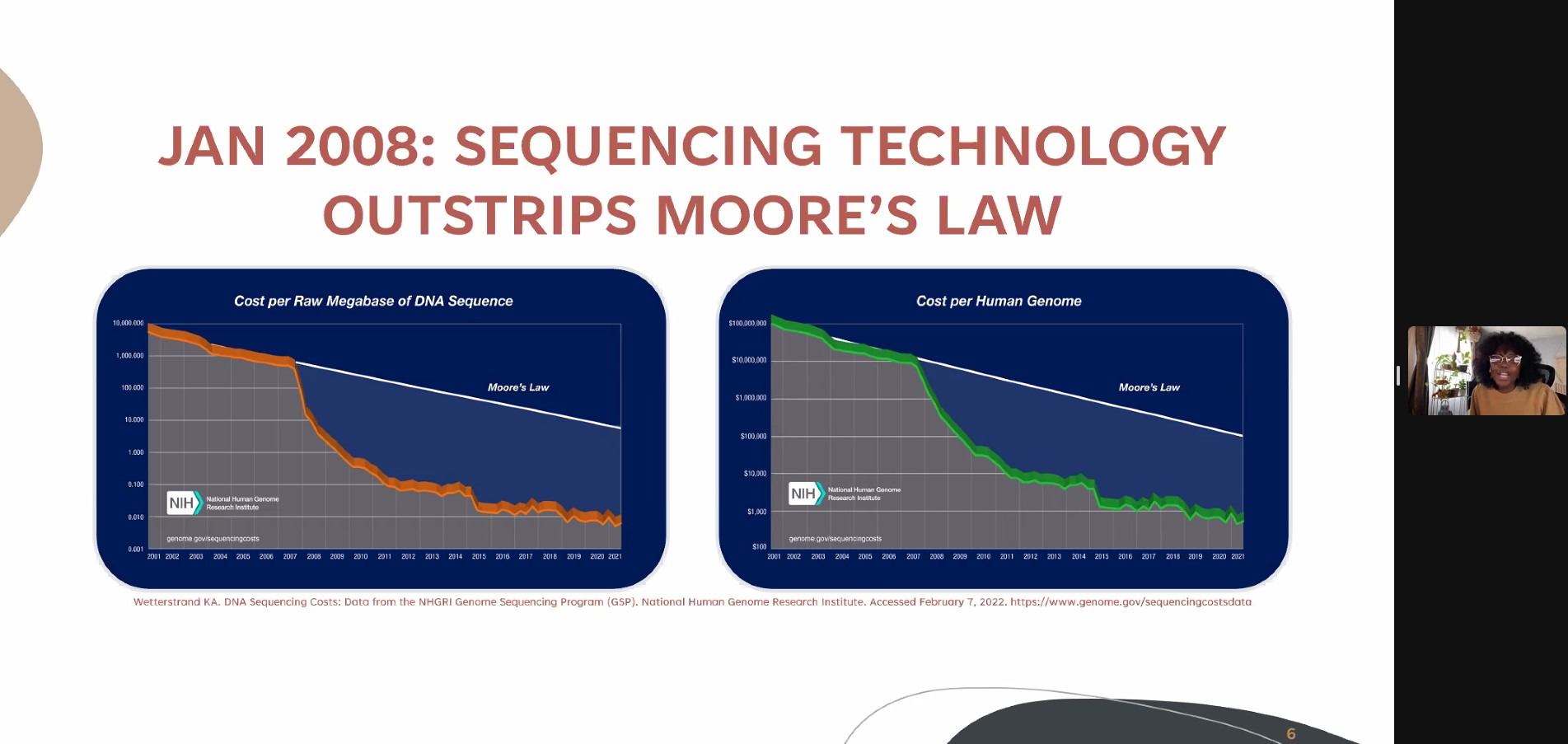
Karyn Mukiri – Predicting the Total Resistome Mugdha Dave – Creation and standardization of automated bacterial pathogen genomic sequencing reporting through informatics
Category: software

Emma J. Griffiths et al. Gigascience. 2022 Feb 16;11:giac003. Background: The Public Health Alliance for Genomic Epidemiology (PHA4GE) (https://pha4ge.org) is a global coalition that is

Jalees Nasir awarded 2021 Fred & Helen Knight Enrichment Award!
Congratulations to PhD student Jalees Nasir, who has been awarded the prestigious 2021 Fred & Helen Knight Enrichment Award, which recognizes exceptional promise in early

Congratulations Emily Panousis on being awarded a 2021 IIDR Summer Student Fellowship! For nine years now, these awards have supported trainees working in IIDR and

Congratulations Martins Oloni on the successful defence of your Masters of Science! New Delhi Metallo-Beta-Lactamase (NDM) plasmid predictions The complete detection and genome sequencing of

A Comparison of Whole Genome Sequencing of SARS-CoV-2 Using Amplicon-Based Sequencing, Random Hexamers, and Bait Capture
Jalees A. Nasir, Robert A. Kozak, Patryk Aftanas, Amogelang R. Raphenya, Kendrick M. Smith, Finlay Maguire, Hassaan Maan, Muhannad Alruwaili, Arinjay Banerjee, Hamza Mbareche, Brian

Nasir, J.A., D.J. Speicher, R.A. Kozak, H.N. Poinar, M.S. Miller, & A.G. McArthur. Preprints 2020, 2020020385. SARS-CoV-2 is a novel betacoronavirus and the aetiological agent
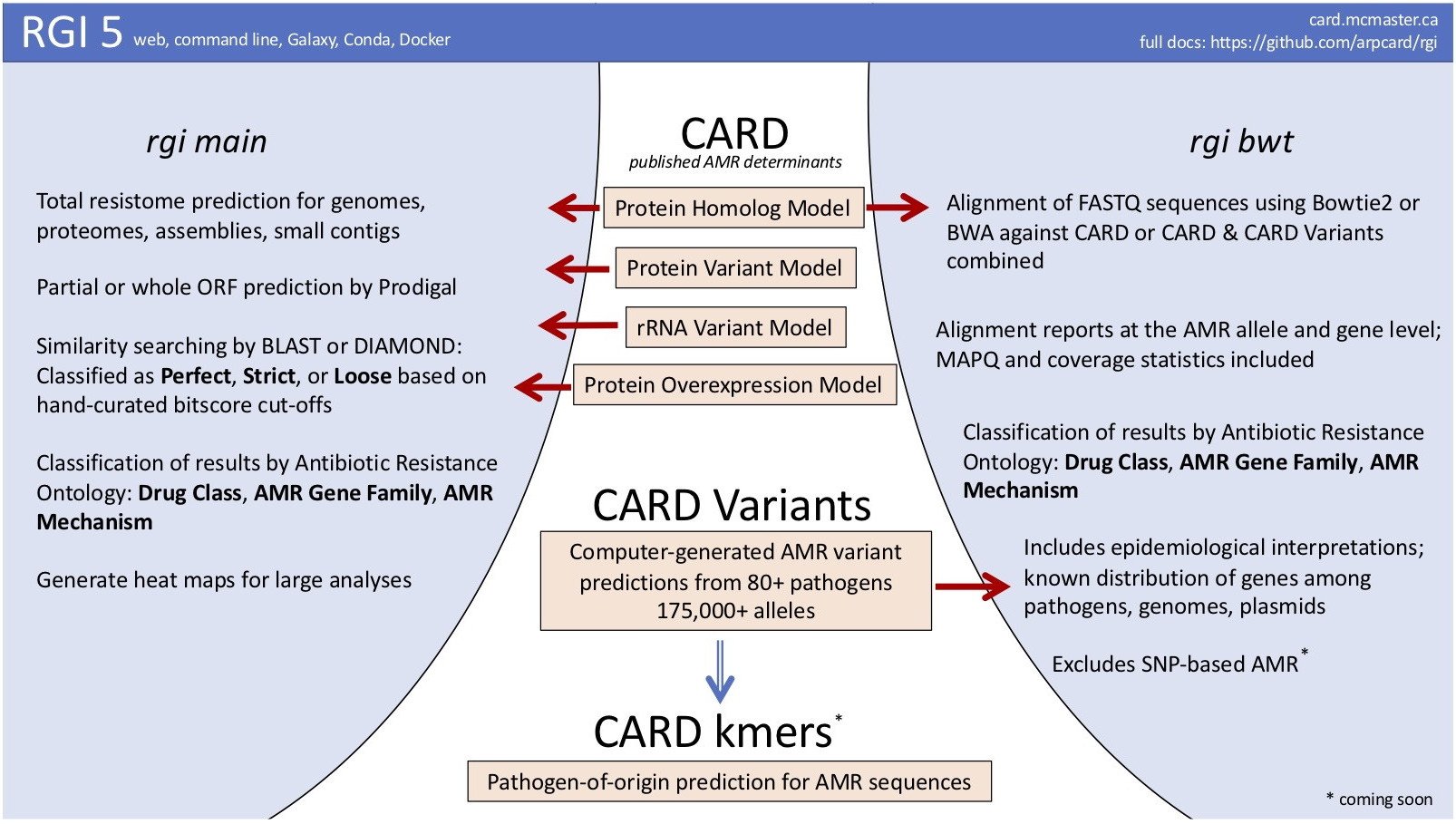
April 2019 big #card_release #RGI5! Resistance Gene Identifier Version 5: entirely new algorithms for metagenomics data, new options for genomes & assemblies. Extensively updated documentation,
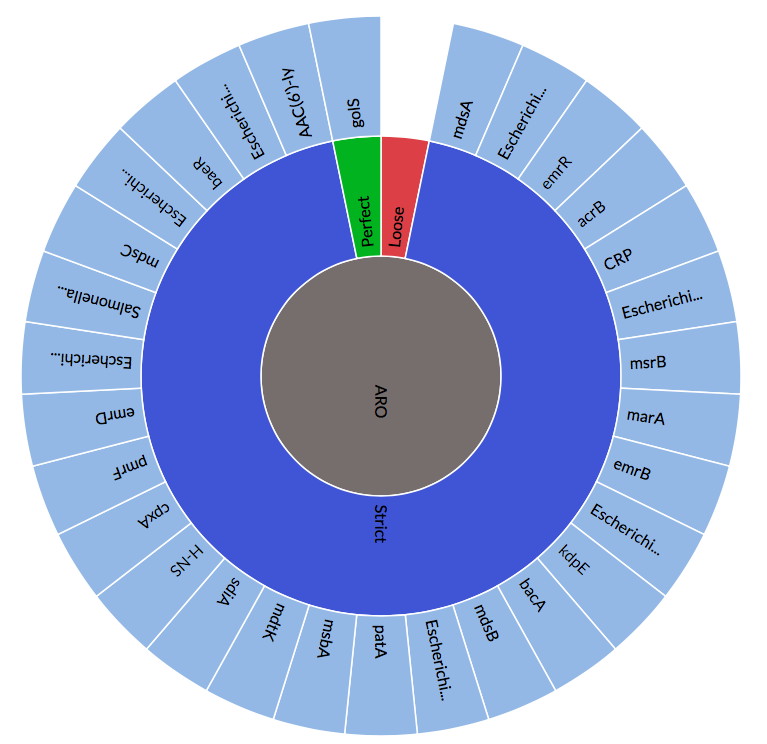
State of the CARD 2019
A week of lectures, demos, and training for the Comprehensive Antibiotic Resistance Database During McMaster Spring Mid-Term Recess (February 18-24), the McArthur lab is pleased

Recent Updates to the Comprehensive Antibiotic Resistance Database
The Comprehensive Antibiotic Resistance Database has been updated, http://card.mcmaster.ca CARD Curation: Addition of HERA, TRU, & ACI beta-lactamases, sul4, and new quinolone efflux pumps. Antibiotic