Databases & Tools
Comprehensive Antibiotic Resistance Database : A bioinformatic database of resistance genes, their products and associated phenotypes. Supported by the Canadian Institutes of Health Research.
Resistance Gene Identifier : Software for prediction of antimicrobial resistance genes and mutations from genomes or metagenomic reads. Supported by the Canadian Institutes of Health Research.
SARS-CoV-2 Illumina GeNome Assembly Line (SIGNAL) : Standardized bioinformatics snakemate workflow for assembly and analysis of SARS-CoV-2 sequencing using Illumina platforms. Development supported by rapid hardware donation from Hewlett Packard Enterprise.
ESKAPE Model, a AI tool purpose-built to identify new antibiotics developed by the Stokes Lab, with interface design by the McArthur Lab. Supported by the Canadian Institutes of Health Research.
Backend processing support provided to the Canadian VirusSeq Data Portal edition of CoVizu (an open source SARS-CoV-2 genome analysis and visualization system). Hardware support from Hewlett Packard Enterprise via Canadian Foundation for Innovation (CFI) funding.
Computing
 The computing environment of the McArthur Lab received financial support from the Canadian Foundation for Innovation, Ontario Research Fund, and Cisco Systems Canada, Inc. We use VMWare Cloud Computing, allowing us to create a wide variety of server, operating system, and software configurations. Our computing resources are part of McMaster’s Service Lab and Repository, hosted by McMaster’s Computer Services Unit. Additional hardware support from Cisco Systems Canada, Inc., Hewlett Packard Enterprise, and the Canadian Foundation for Innovation.
The computing environment of the McArthur Lab received financial support from the Canadian Foundation for Innovation, Ontario Research Fund, and Cisco Systems Canada, Inc. We use VMWare Cloud Computing, allowing us to create a wide variety of server, operating system, and software configurations. Our computing resources are part of McMaster’s Service Lab and Repository, hosted by McMaster’s Computer Services Unit. Additional hardware support from Cisco Systems Canada, Inc., Hewlett Packard Enterprise, and the Canadian Foundation for Innovation.
Git Lab, http://devcard.mcmaster.ca:8888 (requires VPN)
This GitLab server is hosted on in-house CFI-funded research infrastructure, sits behind the McMaster University firewall (requires McMaster virtual private network VPN access), includes daily incremental backup with 45 day retention, and is jointly hosted by the FHS Computer Services Unit and the Laboratory of Dr. Andrew G. McArthur, Department of Biochemistry & Biomedical Sciences, McMaster University. A full description of our infrastructure can be found in the General Repository.
GitLab access is provided for registered users only. Research laboratories in FHS, researchers working in genomics or bioinformatics at McMaster University, or students and faculty using the FHS-CSU computational cluster are welcome to use this GitLab server to support their research programs. Contact mcarthua@mcmaster.ca to register.
Are you a Comprehensive Antibiotic Resistance Database curator or Resistance Gene Identifier tester? This Git Lab server is where you submit and track bug reports, data curation issues, software requests, and your questions.
Are you a user of the FHS-CSU computational cluster? Be sure to read the information available in the General Repository.
McArthur lab members please be sure to read the information available in the McArthurLab Repository.
Comprehensive Antibiotic Resistance Database
All active CARD curators must join the Broad Street Repository. RGI testers are additionally advised to join the RGI Release Repository.
- CARD Production Server (i.e. public website) (http://card.mcmaster.ca)
- CARD Development & Curation server (VPN, http://dune.mcmaster.ca)
Seminars, Journal Clubs, Meet-Ups
 Microbiome Working Group & Human Microbiome Journal Club
Microbiome Working Group & Human Microbiome Journal Club
The Microbiome Working Group is held Tuesdays, 3:30 – 4:30 in HSC 1J9A. Researchers working on microbiome projects are encouraged to bring their laptops and get or give help with software tools, R, experiment planning, or whatever else is going on in your microbiome world. This is a drop-in with no presentations or preparation required.
The Human Microbiome Journal Club is held on the fourth Friday of each month from 3 – 4 in HSC 3N10A. One person chooses a paper to present and discuss. HMJC is typically followed up with Bioinformatics and Beer at the Phoenix. If you would like to present a paper at HMJC, please inform Dr. Jen Stearns at stearns@mcmaster.ca
To get updates about either of these groups (paper topics, cancellations, etc.), please add yourself to the (low-traffic) hmjc-l mailing list here: https://mailman.mcmaster.ca/mailman/listinfo/hmjc-l
Structural Biology Journal Club
The Structural Biology Journal Club is hosted by IIDR principal investigator Dr. Sara Andres (andressn@mcmaster.ca) and is open to all IIDR faculty and trainees. Journal Club meetings are held on the second Wednesday of every month in MDCL 2218.
Bio-Data Lunch
Hosted by the Department of Biology, a Friday lunch to discuss all things biological data. Contact Dr. Ben Bolker for details.
Seminars, Work-in-Progress, Rounds
The Farncombe Family Digestive Health Research Institute has weekly Research in Progress and Noon Rounds.
Seminars! Our favourites are in the Department of Biochemistry & Biomedical Sciences, Department of Biology, MacData Institute, and the Michael G. DeGroote Institute for Infectious Disease Research (IIDR). Watch for the monthly IIDR – Infectious Diseases Joint Rounds!
McArthur Lab Meeting
The McArthur Lab has a group lab meeting most weeks in which we discuss our latest research progress. Thinking about joining the lab? Consider sitting in one of our meetings. Contact mcarthua@mcmaster.ca for details.
Suggested Reading & Online Tutorials
Drug Resistance
- Wright & Brown: The greatest threat of our time — antibiotic resistance (National Post 2015)
- 2019 When Antibiotics Fail: The Expert Panel on the Potential Socio-Economic Impacts of Antimicrobial Resistance in Canada
- 2019 CDC AR Threats Report [USA]
- 2018 Progress Report on the 2015 Federal Action Plan on Antimicrobial Resistance and Use [Canada]
- 2017 Tackling Antimicrobial Resistance and Antimicrobial Use: A Pan-Canadian Framework for Action
- 2016 Forum on Genomics and Antimicrobial Resistance – Workshop Report [Canada]
- 2016 UK Review on Antimicrobial Resistance
- 2015 Federal Action Plan on Antimicrobial Resistance and Use in Canada: Building on the Federal Framework for Action
- 2015 Reports of the Auditor General of Canada Report — Antimicrobial Resistance
- 2014 U.S. National Strategy for Combating Antibiotic-Resistant Bacteria
- 2014 Antimicrobial Resistance and Use in Canada: A Federal Framework for Action
- 2014 WHO Antimicrobial Resistance Global Report on Surveillance
Bioinformatics, Computing, Genome Science
- Bioinformatics Goes Back to the Future [Nature]
- Developing Bioinformatics and Programming Skills
- Pick Up Python [Nature]
- Introduction to Shell (command line training)
- Free MIT video lecture series: Introduction to Python
- Rosalind, online bioinformatics problem sets
- Coursera, online courses
- DataCamp, teaches basics of Python and R (tutorials target the Data Sciences)
- Codeacademy, learn coding basics, a good starting point for Python
- Suggested books: Bioinformatics Data Skills, Practical Computing for Biologists
- WikiBooks: Next Generation Sequencing (NGS)
- ISB Biocuration Training Materials
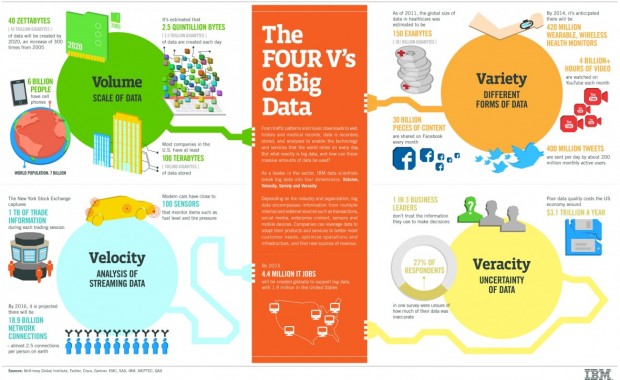
- The Four Vs of Big Data
- Internet of DNA [MIT Tech Review]
- Craft beautiful equations in Word with LaTeX
Science in Society, Women in Science, Social Justice
- Science Culture: Where Canada Stands
- Reflecting on Fifty Years of Progress for Women in Science [DNA and Cell Biology]
- The Henrietta Lacks Legacy Grows [dig deeper: The Immortal Life of Henrietta Lacks]
- The Man Behind the Machine [Nature] [dig deeper: Alan Turing: The Enigma]