Today the Comprehensive Antibiotic Resistance Database turns 10 years old! While it started a few years before this in the laboratory of Dr. Gerry Wright,
Category: news

Brian Alcock has received a Gradaute Student Association Award for Contributions by Non-Academic Staff! Brian works as CARD’s Lead Curator but also has trained countless

Two current lab members are changing their roles, with Mugdha Dave starting her MSc studies and Dr. Sheridan Baker transitioning from CanCOGeN work to a

Today we say goodbye to William Huynh, who first joined us as an undergraduate thesis student and then stayed on to work as the Comprehensive

Jalees Nasir awarded 2021 Fred & Helen Knight Enrichment Award!
Congratulations to PhD student Jalees Nasir, who has been awarded the prestigious 2021 Fred & Helen Knight Enrichment Award, which recognizes exceptional promise in early

It is with great pleasure the McArthur lab announces that Lead Developer for the Comprehensive Antibiotic Resistance Database (CARD), Amos Raphenya, is starting his M.Sc.

McMaster is launching The Global Nexus for Pandemics and Biological Threats, to ensure Canada and the world are better able to manage the human and
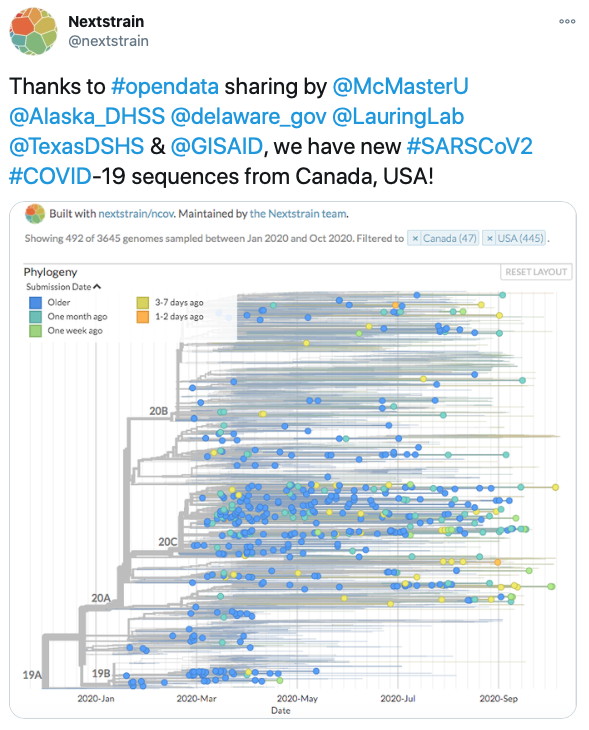
McArthur Lab & CARD during SARS-CoV-2
Quick update on the status of the McArthur Lab and the Comprehensive Antibiotic Resistance Database. While our home institution McMaster University is closed to on-site

(left to right) Back row: Amos Raphenya, Arman Edalatmand, Brian Alcock, Andrew McArthur, David Speicher, Martins Oloni, Sohaib Syed. Front row: William Huynh, Anna-Lisa Nguyen,

Jalees Nasir transferred from the McMaster Biochemistry & Biomedical Sciences Masters program to the Ph.D. program today! Jalees’s work focusses on the developing new genomic

David Braley Centre for Antibiotic Discovery gives researchers a fighting chance against antimicrobial resistance. A forward-looking McMaster donor is investing $7 million in a new

Congratulations to Kara Tsang for winning the 2018 Michael G. DeGroote Institute for Infectious Disease Research (IIDR) Michael Kamin Hart Memorial Scholarship (MSc), the highest academic

Dr. McArthur has been busy doing some Government outreach. In early 2018 he was a Panellist on Artificial Intelligence in Healthcare at Norwegian Health Ministry &
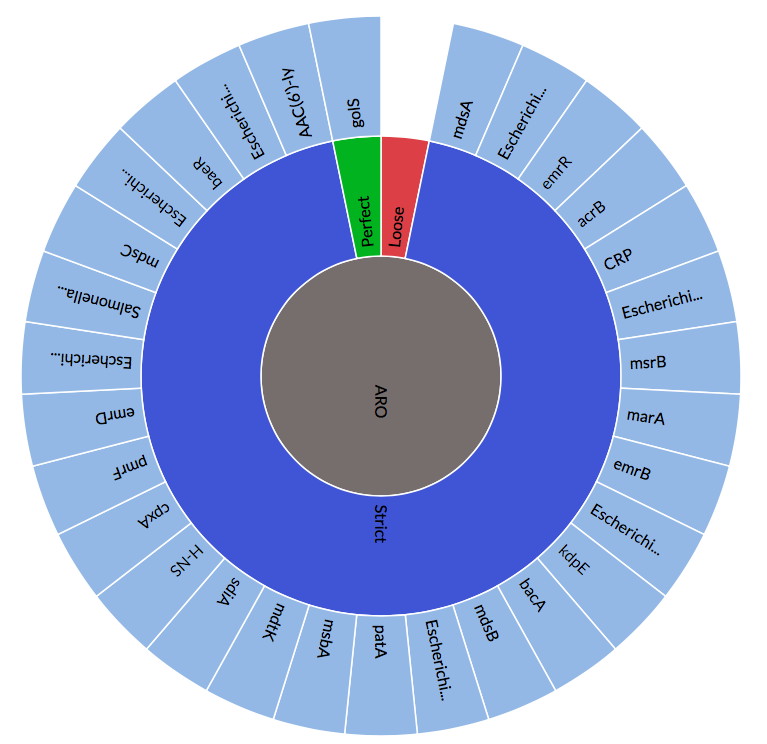
Recent Updates to the Comprehensive Antibiotic Resistance Database
The Comprehensive Antibiotic Resistance Database has been updated, http://card.mcmaster.ca CARD Curation: Addition of HERA, TRU, & ACI beta-lactamases, sul4, and new quinolone efflux pumps. Antibiotic
Major Update of the Comprehensive Antibiotic Resistance Database
The Comprehensive Antibiotic Resistance Database has been updated, http://card.mcmaster.ca This February 2018 release is our largest to date and includes new data types, a new

Congratulations to our 3rd year Biochemistry & Biomedical Sciences students upon completion of their research projects! Both will be staying on as summer research students.

Congratulations to 4th Year Biomedical Discovery & Commercialization student and McArthur lab member Suman Virdee for her CIHR Undergraduate Summer Studentship Award for her summer