Wlodarski, M.A., T.T.Y. Lau, B.P. Alcock, A.R. Raphenya, T.E. Ta, F. Maguire, R.G. Beiko, & A.G. McArthur. 2025. Unmasking antibiotic resistance genes and their pathogen
Category: bioinformatics

Characterizing the Influence of Dietary Fibre Consumption on the Composition and Function of Fibre Fermenting Bacteria in the Infant Gut Microbiome By COLIN Y. BRUCE,

Baker et al. 2025. Journal of the Association of Medical Microbiology and Infectious Disease Canada, in press. Background: Clostridioides difficile is a bacillus that can

33rd Conference on Intelligent Systems for Molecular Biology
Mukiri, K., B. Alcock, A. Raphenya, & A.G. McArthur. 2025. Mind the gap: predicting the total bacterial resistome in the fight against antimicrobial resistance. Poster

It is with great pleasure that we announce that Dr. Jalees Nasir has been awarded the 2025 McMaster Health Sciences Graduate Student Association (HSGSA) Impact

Lu et al. Genome Med. 2025 May 6;17(1):46. Background: Antimicrobial resistant (AMR) pathogens represent urgent threats to human health, and their surveillance is of paramount

Antibiotics and Resistance: Past, Present, and Future By DIRK L HACKENBERGER, B.Sc. A thesis submitted to the school of graduate studies in partial fulfillment of

Congratulations Dr. Jalees Nasir!
Designing Molecular Fishhooks for Virus Survellance Platforms By JALEES A. NASIR, B.Sc A Thesis Submitted to the School of Graduate Studies in partial fulfilment of

Wlodarski, M.A., T.T.Y. Lau, B.P. Alcock, A.R. Raphenya, F. Maguire, R.G. Beiko, T.E. Ta, & A.G. McArthur. 2025. Unmasking antibiotic resistance genes and their pathogen

Hackenberger et al. Appl Environ Microbiol. 2025 Feb 28:e0187624. Better interrogation of antimicrobial resistance requires new approaches to detect the associated genes in metagenomic samples.

Two alumni of the McMaster Biochemistry graduate program have joined the McArthur Lab! Dr. Emily Bordeleau got her PhD from McMaster in 2022 under the
Nasir et al. NAR Genomics & Bioinformatics. 2024 Dec 18;6(4):lqae176. The incorporation of sequencing technologies in frontline and public health healthcare settings was vital in
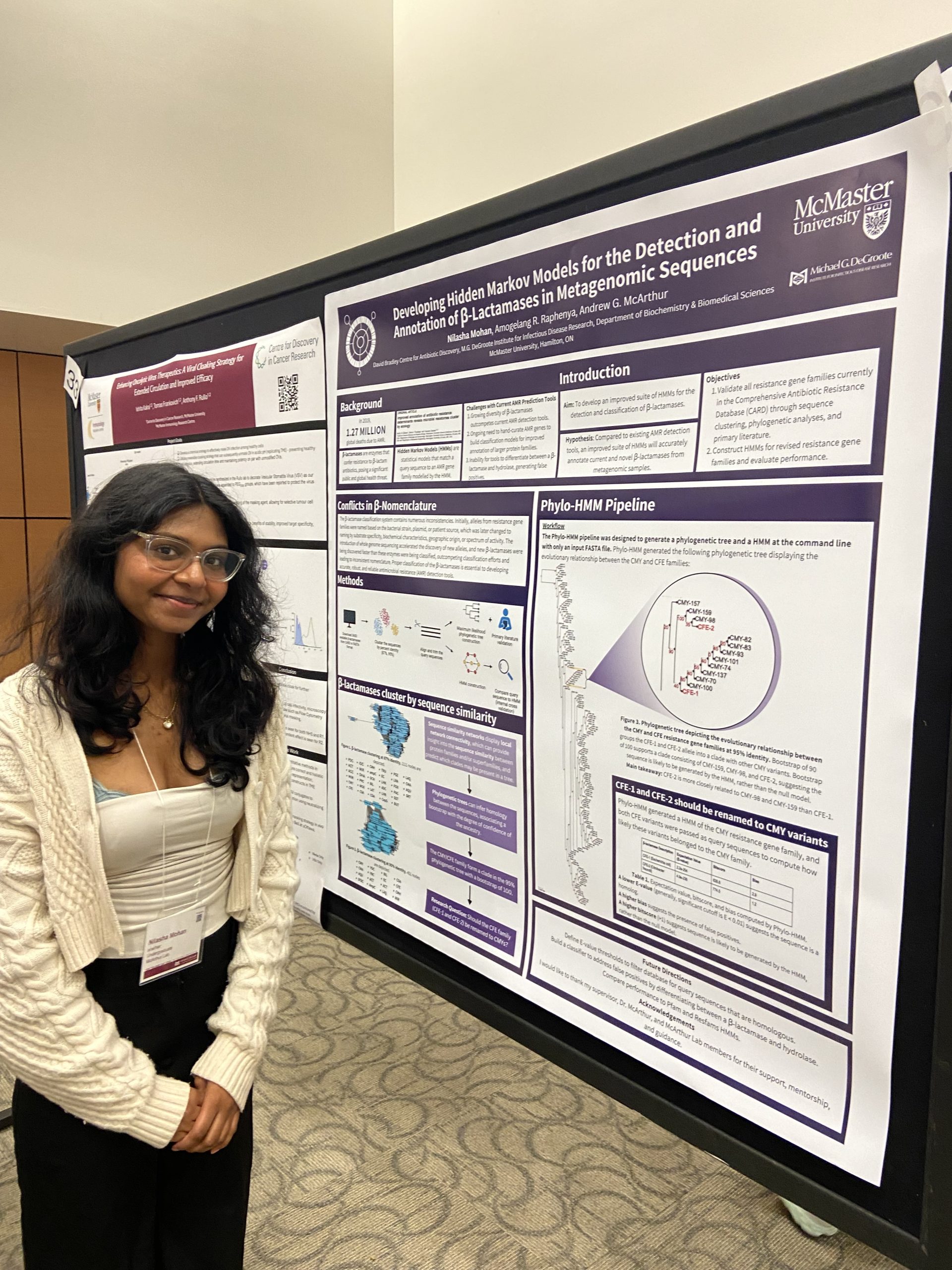
IIDR Trainee Day 2024!
A great day of posters and presentations by the trainees of the Michael G. DeGroote Institute of Infectious Disease Research! Nilasha Mohan, Undergraduate student. Developing

Gill et al. 2024. The Canadian VirusSeq Data Portal & Duotang: open resources for SARS-CoV-2 viral sequences and genomic epidemiology. Microb Genom. 2024 Oct;10(10):001293. The
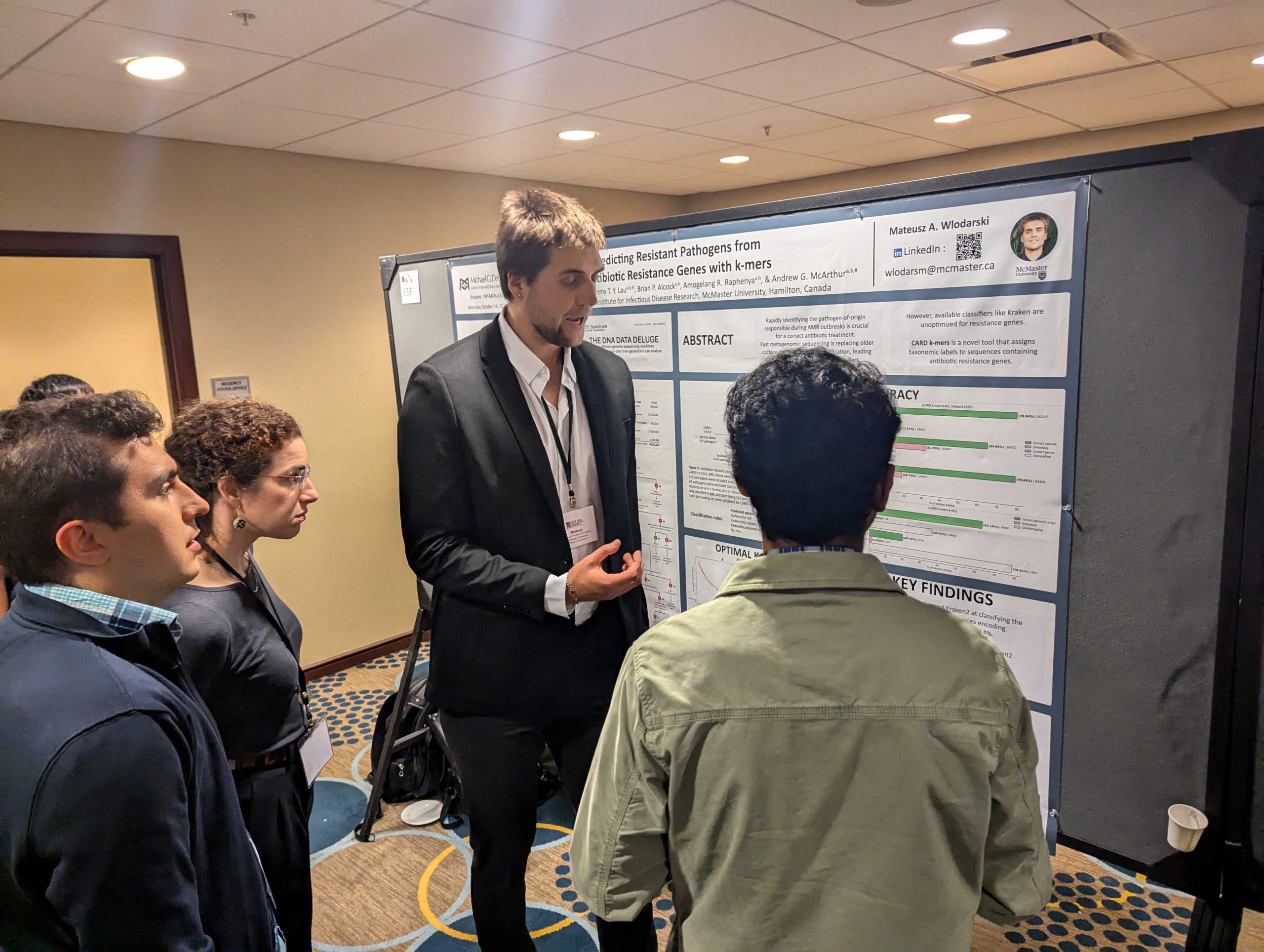
Wlodarski, M.A., T.T.Y. Lau, A.R. Raphenya, B.P. Alcock, & A.G. McArthur. 2024. Accurate pathogen-of-origin classification of antibiotic resistance genes with CARD k-mers. Presentation at the

This one-hour webinar, entitled “DNA sequencing in infectious disease surveillance, diagnosis, and management,” features talks from Andrew McArthur (Professor, Biochemistry and Biomedical Sciences) and Rubayet

Gill, E.E., B. Jia, C.L. Murall, R. Poujol, M.Z. Anwar, N.S. John, J. Richardsson, A. Hobb, A.S. Olabode, A. Lepsa, A.T. Duggan, A.D. Tyler, A.

Alcock, B.P., E.A. Bordeleau, & A.G McArthur. 2024. Improving aminoglycoside resistance surveillance and stewardship efforts through nomenclature harmonization with the Comprehensive Antibiotic Resistance Database. Presentation

Antimicrobial Resistance – Genomes, Big Data and Emerging Technologies – Wellcome Trust, UK
Mukiri, K.M., B.P. Alcock, & A.G. McArthur. 2024. Increasing the predictive accuracy of the Resistance Gene Identifier by abandoning sole reliance on bitscore. Ta, T.E.,

