April 2019 big #card_release #RGI5! Resistance Gene Identifier Version 5: entirely new algorithms for metagenomics data, new options for genomes & assemblies. Extensively updated documentation,
Category: bioinformatics
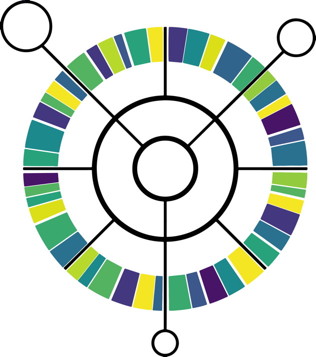
The Comprehensive Antibiotic Resistance Database has been updated, http://card.mcmaster.ca CARD Curation: Expanded MCR, OXA & IMP beta-lactamase, and macrolide phosphotransferase (MPH) sequence curation. Updated nomenclature
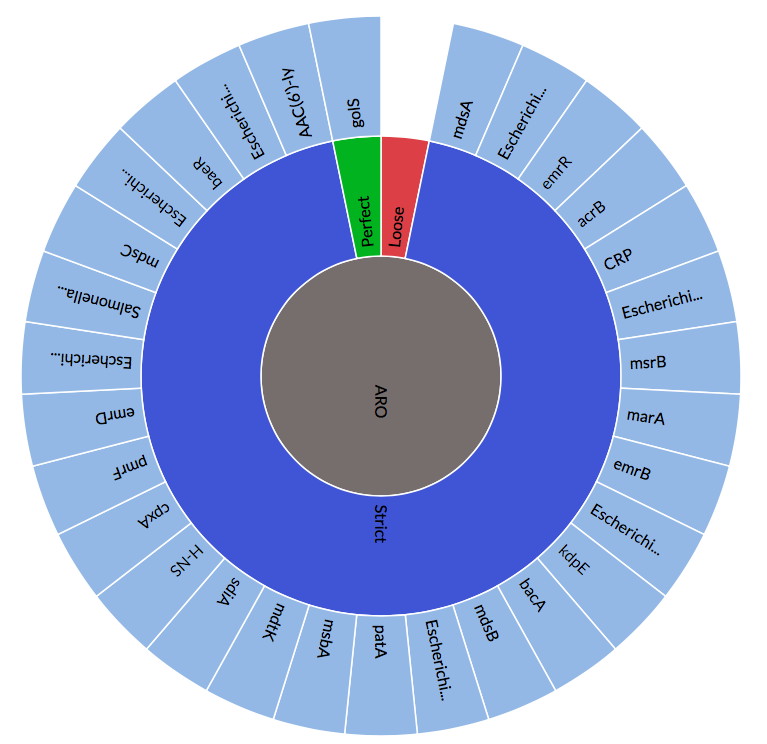
State of the CARD 2019
A week of lectures, demos, and training for the Comprehensive Antibiotic Resistance Database During McMaster Spring Mid-Term Recess (February 18-24), the McArthur lab is pleased

Antimicrobial Resistance: Emergence, Transmission, and Ecology (ARETE). R. Beiko (PI; Dalhousie University), F. Brinkman (co-PI, Simon Fraser University), A.G. McArthur (co-Applicant) + 4 additional co-Applicants.
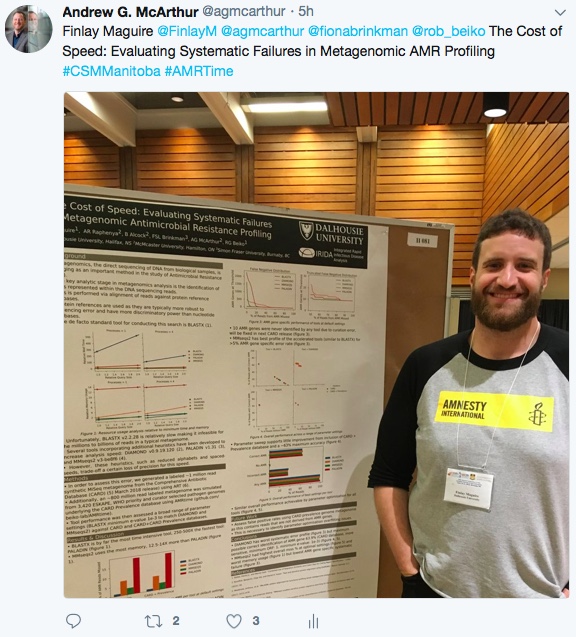
Maguire, F., B. Alcock, F.S. Brinkman, A.G. McArthur, & R.G. Beiko. 2018. AMRtime: Rapid Accurate Identification of Antimicrobial Resistance Determinants from Metagenomic Data. Oral presentation
Welcome #TeamVirulence, left to right: Rachel Tran (Biochem 3R06), Sally Min (BiomedDC 4A15), Anatoly Miroshnichencko (BiomedDC 4A15), and Rafik El Werfalli (BiomedDC 4A15), who are

Tsang, K.K., H. Zubyk, S. Chou, G.D. Wright, & A.G. McArthur. Decoding bad bags: Predicting antibiotic resistance phenotypes from genotype. Oral presentation at the Canadian Society of
Alcock, B., A.R. Raphenya, A.N. Sharma, K.K. Tsang, T.T.Y. Lau, A. Hernandez-Koutoucheva, & A.G. McArthur. 2018. Data and curation in the Comprehensive Antibiotic Resistance Database.
Kara Tsang passed her graduate transfer exam today, officially moving from the McMaster Biochemistry & Biomedical Sciences Masters program to the Ph.D. program. Kara’s work

Tammy Lau has been awarded a prestigious Michael G. DeGroote Institute for Infectious Disease Research (IIDR) Summer Student Fellowship for her work on development of k-mer approaches

Recent Updates to the Comprehensive Antibiotic Resistance Database
The Comprehensive Antibiotic Resistance Database has been updated, http://card.mcmaster.ca CARD Curation: Addition of HERA, TRU, & ACI beta-lactamases, sul4, and new quinolone efflux pumps. Antibiotic

The Comprehensive Antibiotic Resistance Database: Expanded tools for molecular surveillance of antimicrobial resistance (AMR) in the environment, agriculture, and clinic. A.G. McArthur (PI), G. Van

The McArthur lab and the Comprehensive Antibiotic Resistance Database are proud to join the Canadian Anti-Infective Innovation Network, International Genomic Epidemiology Application Ontology Consortium, and Integrated Rapid

Major Update of the Comprehensive Antibiotic Resistance Database
The Comprehensive Antibiotic Resistance Database has been updated, http://card.mcmaster.ca This February 2018 release is our largest to date and includes new data types, a new

Tsang, K.K. & A.G. McArthur. 2017. Encoding the efflux pump phenomena. Oral presentation at the Second American Society for Microbiology Meeting on Rapid Applied Microbial
Building upon her successful Biochem 3A03 project, Tammy Lau is staying in the lab for 2017-2018 as part of her Biochem 4T15 Research Thesis. Tammy’s

Today we say farewell to Arjun Sharma & Suman Virdee. Arjun joined the lab as a second year volunteer, staying to perform a Biochem 3R06

We haven’t been travelling much this year, but our collaborators have been busy! Dearborn, D.C., A.B. Gager, A.G. McArthur, M.E. Gilmour, E. Mandzhukova, R.A. Mauck.

Congratulations to this year’s crop of BiomedDC 4A15 thesis students for 8 month research projects well done! From left to right: Suman Virdee – Developing a

