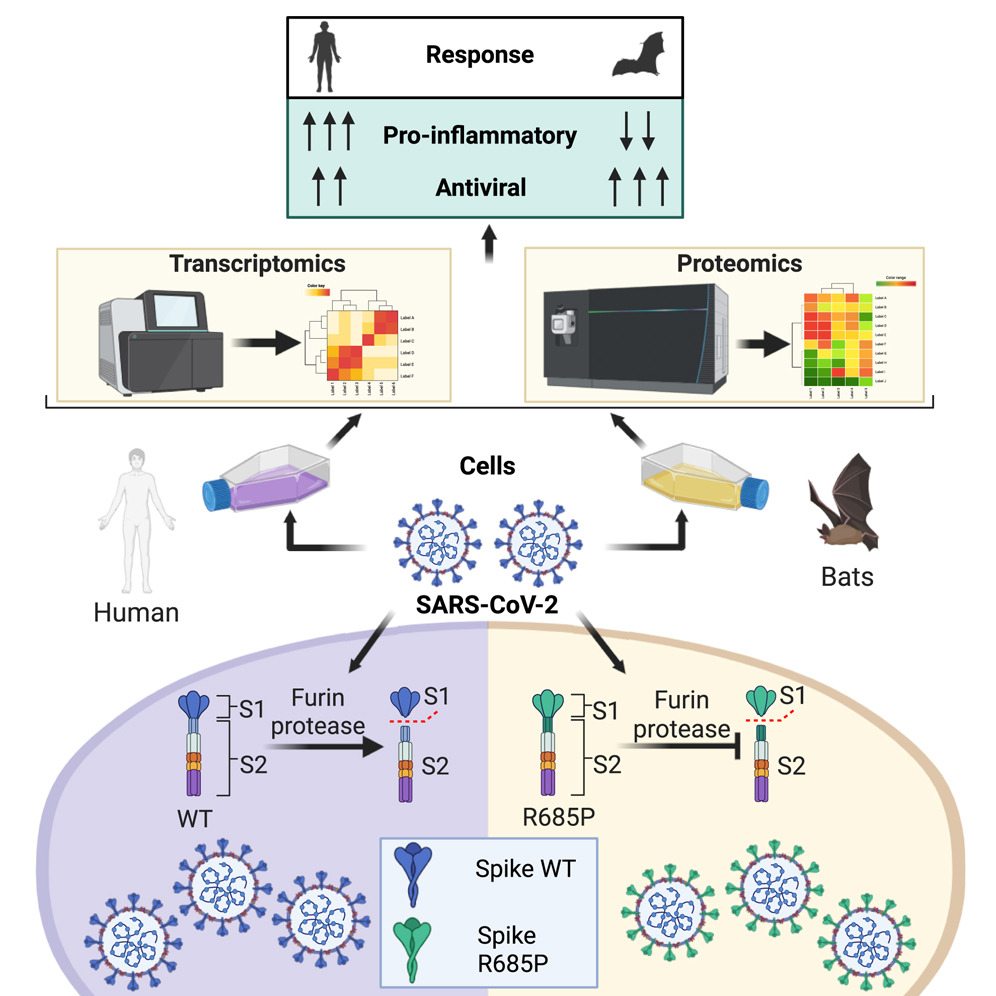
Baid et al. 2015. Cell Reports 44, 115929. SARS-CoV-2 has caused the largest known coronavirus pandemic and is believed to have emerged from insectivorous bats.
Category: SARS-CoV-2

Baker et al. IDCases. 2025 May 28:40:e02274. Background: Differentiating severe systemic inflammatory syndromes from sepsis can be challenging. The diagnostic process may be further complicated

It is with great pleasure that we announce that Dr. Jalees Nasir has been awarded the 2025 McMaster Health Sciences Graduate Student Association (HSGSA) Impact

Congratulations Dr. Jalees Nasir!
Designing Molecular Fishhooks for Virus Survellance Platforms By JALEES A. NASIR, B.Sc A Thesis Submitted to the School of Graduate Studies in partial fulfilment of

This week we say farewell to Dr. Sheridan Baker, who joined us in early 2021 as lead molecular biologist for our Ontario Genomics Coalition (ONCoV)

Nirmalarajah et al. BMC Infect Dis. 2025 Jan 28;25(1):132. Background: Drivers of COVID-19 severity are multifactorial and include multidimensional and potentially interacting factors encompassing viral
Nasir et al. NAR Genomics & Bioinformatics. 2024 Dec 18;6(4):lqae176. The incorporation of sequencing technologies in frontline and public health healthcare settings was vital in

Gill et al. 2024. The Canadian VirusSeq Data Portal & Duotang: open resources for SARS-CoV-2 viral sequences and genomic epidemiology. Microb Genom. 2024 Oct;10(10):001293. The

CSM Mukiri, K.M., B.P. Alcock, & A.G. McArthur. 2024. Increasing the predictive accuracy of the Resistance Gene Identifier by abandoning sole reliance on bitscore.
Chronic COVID-19 infection in an immunosuppressed patient shows changes in lineage over time: a case report
Sheridan J C Baker , Landry E Nfonsam , Daniela Leto , Candy Rutherford , Marek Smieja , & Andrew G McArthur Virol J. 2024

Baker, S.J.C., J. Maciejewski, M.-T. Usuanlele, J. Gilchrist, D.R. Sharma, D. Bulir, M. Smieja, M. Loeb, M.G. Surette, A.G. McArthur, & D. Mertz. 2023. Investigating

Jacob et al. Emerg Infect Dis. 2023 Jun 12;29(7). Isolating and characterizing emerging SARS-CoV-2 variants is key to understanding virus pathogenesis. In this study, we

The SARS-CoV-2 Illumina GeNome Assembly Line (SIGNAL) has been updated to version 1.6.2, with an important hotfix to address software version incompatibility for rules using

Tsang et al. Microbiol Spectr. 2023 Apr 24;e0190022. Genomic epidemiology can facilitate an understanding of evolutionary history and transmission dynamics of a severe acute respiratory

Baker, S.J.C., J. Maciejewski, M.-T. Usuanlele, J. Gilchrist, D.R. Sharma, D. Bulir, M. Smieja, M. Loeb, M.G. Surette, AG. McArthur, & D. Mertz. 2023. Investigating

Congratulations Ph.D. student Jalees Nasir!
Congratulations Jalees Nasir on being awarded the Dr. Jordon Page Harshman Bursary! The Dr. Jordan Page Harsham Bursary was established in 2011 by the Harshman

Congratulations Dr. Sheridan Baker on being awarded a 2-year Mitacs Elevate Postdoctoral Fellowship, which will allow him to train jointly in clinical epidemiology, molecular epidemiology,

Summer Conferences 2022
(presenters in bold; lab members underlined) Baker, S.J.C., S. Yaqoob, M.-T. Usuanlele, J. Gilchrist, M. Smieja, M. Loeb, S. Khan, J. Pernica, M.G. Surette, A.G.

The Dataharmonizer: a Tool for Faster Data Harmonization, Validation, Aggregation, and Analysis of Pathogen Genomics Contextual Information
Gill et al. 2022. Preprints 10.20944/preprints202206.0335.v1 Pathogen genomics is a critical tool for public health surveillance, infection control, outbreak investigations, as well as research. In

Emma J. Griffiths et al. Gigascience. 2022 Feb 16;11:giac003. Background: The Public Health Alliance for Genomic Epidemiology (PHA4GE) (https://pha4ge.org) is a global coalition that is