Jalees A. Nasir, Robert A. Kozak, Patryk Aftanas, Amogelang R. Raphenya, Kendrick M. Smith, Finlay Maguire, Hassaan Maan, Muhannad Alruwaili, Arinjay Banerjee, Hamza Mbareche, Brian
Category: genomics
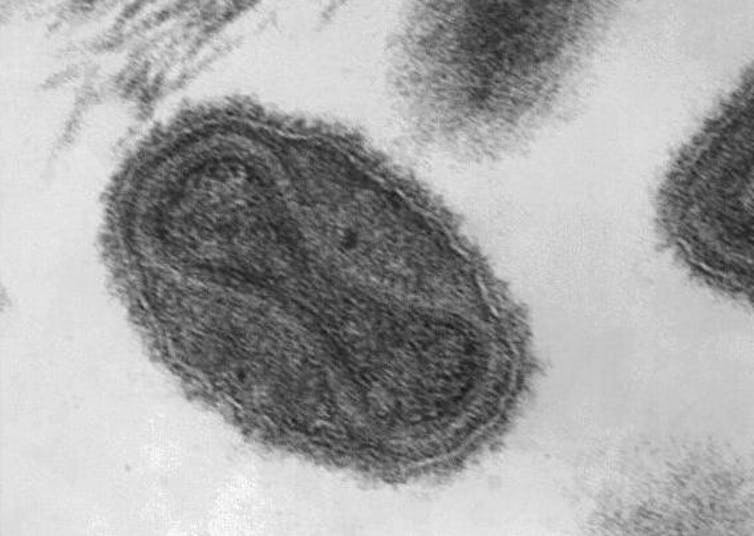
Ana T Duggan, Jennifer Klunk, Ashleigh F Porter, Anna N Dhody, Robert Hicks, Geoffrey L Smith, Margaret Humphreys, Andrea M McCollum, Whitni B Davidson, Kimberly
Maan, H., H. Mbareche, A.R. Raphenya, A. Banerjee, J.A. Nasir, R.A. Kozak, N. Knox, S. Mubareka, A.G. McArthur, & B. Wang. The Lancet Digital Health,

Thanks to hard work by Jalees Nasir, Amos Raphenya, Dr. Kendrick Smith (Perimeter Institute), and Dr. Finlay Maguire (Dalhousie) with help from our Ontario Coronavirus

Krishna A Srinivasan, Suman K Virdee, Andrew G McArthur Brief Funct Genomics 2020 May 16 [Epub ahead of print] RNA sequencing (RNA-Seq) is a complicated

More information: The IIDR & DBCAD Announce Funding for Collaborative Projects

New CARD Team Member!
The McArthur lab welcomes new Curator-Developer William Huynh to the Comprehensive Antibiotic Resistance Database staff. Looking forward to pushing CARD forward with William’s help!

The McArthur lab is honoured to collaborate with our clinical colleagues across Ontario in sequencing of SARS-CoV-2 clinical isolates, to better understand the epidemiology of

The McArthur lab welcomes back summer students Rachel Tran & Arman Edalatmand plus first timers Marcel Jansen & Emily Panousis! These four will be covering

Welcome Ashraf Bazan, who has joined the lab and the McMaster Biochemistry & Biomedical Sciences graduate program! A familiar face in the IIDR as he started

Banerjee, A., J.A. Nasir, P. Budylowski, L. Yip, P. Aftanas, N. Christie, A. Ghalami, K. Baid, A.R. Raphenya, J.A. Hirota, M.S. Miller, A.J. McGeer, M.A.
Drs. Andrew McArthur & Arinjay Banerjee giving the background story on isolation of the live SARS-CoV-2 virus in Canada.

Congratulations finishing thesis students!
COVID19 kept us from celebrating together, but we are very proud of the work you have accomplished! Rachel Tran | Biochem 4T15 – Exploring the
McArthur Lab & CARD during SARS-CoV-2
Quick update on the status of the McArthur Lab and the Comprehensive Antibiotic Resistance Database. While our home institution McMaster University is closed to on-site

Nasir, J.A., D.J. Speicher, R.A. Kozak, H.N. Poinar, M.S. Miller, & A.G. McArthur. Preprints 2020, 2020020385. SARS-CoV-2 is a novel betacoronavirus and the aetiological agent

CARD 2020: antibiotic resistome surveillance with the Comprehensive Antibiotic Resistance Database
Alcock BP, Raphenya AR, Lau TTY, Tsang KK, Bouchard M, Edalatmand A, Huynh W, Nguyen A-LV, Cheng AA, Liu S, Min SY, Miroshnichenko A, Tran

Guitor AK, Raphenya AR, Klunk J, Kuch M, Alcock B, Surette MG, McArthur AG, Poinar HN, Wright GD. Editor’s Pick! Antimicrobial Agents and Chemotherapy 2019 Oct

Waglechner N, McArthur AG, Wright GD. Nat Microbiol. 2019 Aug 12. [Epub ahead of print] Glycopeptide antibiotics are produced by Actinobacteria through biosynthetic gene clusters

Empowering conventional antibiotics with new drug leads targeting the outer-membrane of Gram-negative pathogens. E. Brown (PI), G.D. Wright (co-I), B. Coombes (co-I), A.G. McArthur (co-I),
MacData Summer School 2019
Dr. McArthur and PhD student Kara Tsang taught together at the 2019 MacData Institute Summer School, with Dr. McArthur reviewing biocuration and bioinformatics for genomic
