Baker, S.J.C., J. Maciejewski, M.-T. Usuanlele, J. Gilchrist, D.R. Sharma, D. Bulir, M. Smieja, M. Loeb, M.G. Surette, A.G. McArthur, & D. Mertz. 2023. Investigating
Category: genomics

Jacob et al. Emerg Infect Dis. 2023 Jun 12;29(7). Isolating and characterizing emerging SARS-CoV-2 variants is key to understanding virus pathogenesis. In this study, we

While many know of Amogelang (Amos) Raphenya as the Lead CARD Developer and a local, national, and international resource for genomic surveillance of antimicrobial resistance,

Colin Bruce joins us by way of the laboratory of Dr. Jennifer Stearns and will be completing the last year of his PhD studies with

Today the Comprehensive Antibiotic Resistance Database turns 10 years old! While it started a few years before this in the laboratory of Dr. Gerry Wright,

CARD and CZ ID are thrilled to launch a new CZ ID module that allows researchers to detect and analyze antimicrobial resistance (AMR) genes in

The SARS-CoV-2 Illumina GeNome Assembly Line (SIGNAL) has been updated to version 1.6.2, with an important hotfix to address software version incompatibility for rules using

Tsang et al. Microbiol Spectr. 2023 Apr 24;e0190022. Genomic epidemiology can facilitate an understanding of evolutionary history and transmission dynamics of a severe acute respiratory

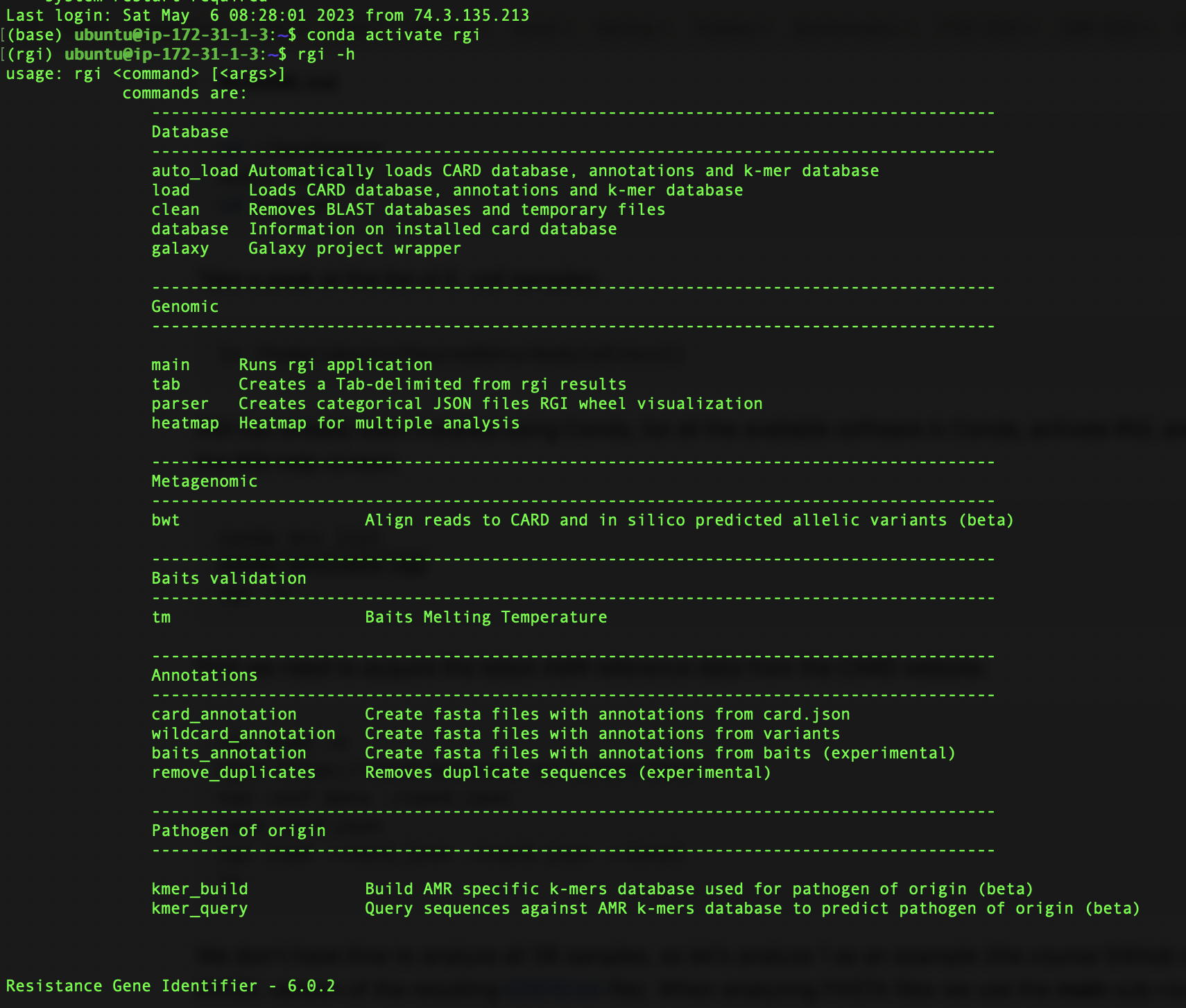
Alcock, B.P., A.R. Raphenya, A. Edalatmand, & A.G. McArthur. 2023. The Comprehensive Antibiotic Resistance Database – curating the global resistome. Oral presentation at the 16th
CBW & VTEC 2023
They’re back! Thanks to Karyn Mukiri, Jalees Nasir, and Madeline McCarthy for serving as Teaching Assistants for the re-booted Canadian Bioinformatics Workshop in Infectious Disease
