This month we say farewell to Briony Lago as she returns to her undergraduate studies in Chemical Biology at McMaster. Briony joined us as part
Category: bioinformatics
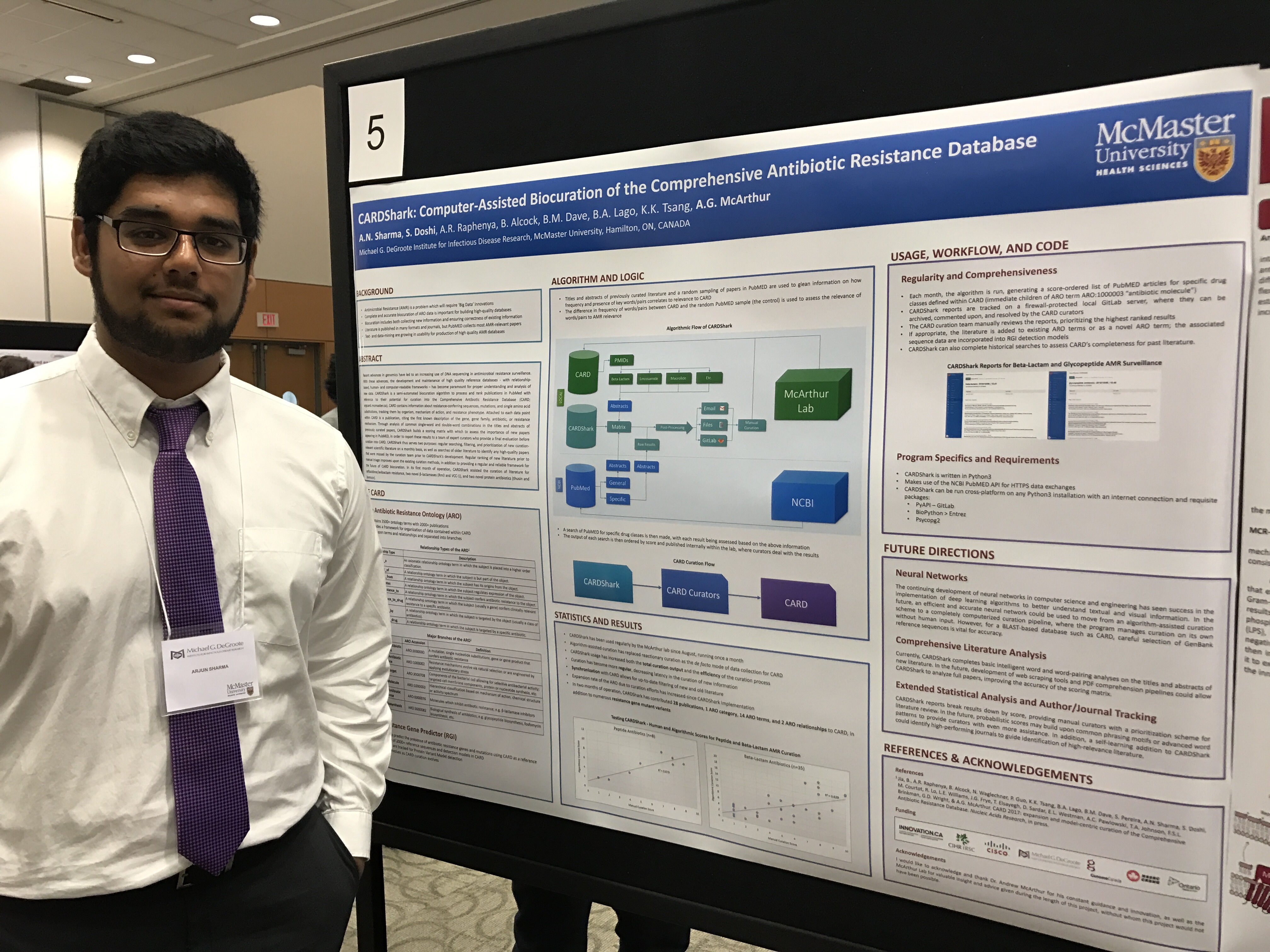
IIDR Trainee Day & USRA Poster Session 2016
Three of our undergraduate students gave poster presentations in the last month. Arjun Sharma (Biochemistry & Biomedical Sciences 3rd year) outlined his work on developing the

Ni X, Davis JH, Jain N, Razi A, Benlekbir S, McArthur AG, Rubinstein JL, Britton RA, Williamson JR, Ortega J. Nucleic Acids Res. 2016 Sep

A cross-national research consortia co-led by McMaster’s Andrew McArthur is receiving two of 16 federal grants to further develop a big data solution to the

Kara Tsang (graduate, McMaster BDC) – Kara did her BiomedDC 4A15 thesis in the lab, focused on Pseudomonas resistome prediction and cystic fibrosis-associated metagenomic sequencing

McArthur, A.G., B. Jia, A.R. Raphenya, P. Guo, K. Tsang, B. Dave, B. Alcock, B. Lago, N. Waglechner, & G.D. Wright. 2016. The Comprehensive Antibiotic Resistance

Congratulations to Kara Tsang and Zachary Lin on completion of their Biomedical Discovery and Commercialization (BDC) 4A15 thesis research! Both Kara & Zachary presented their research

Combatting Antibiotic Resistance Using Surveillance – click on the image to watch the 10 minute video. More details here.

Brian Alcock has joined the McArthur Lab to lead curation of the Comprehensive Antibiotic Resistance Database (arpcard.mcmaster.ca). Brian recently completed his MSc in the laboratory

The Inaugural Biomedical Discovery & Commercialization (BDC) Symposium
Zachary Lin – Adapting Galaxy bioinformatics to outbreak-associated Clostridium difficile The completion of the human genome project in 2001 sparked the beginning of a sequencing revolution

Dr. McArthur gave a MacTalk at McMaster’s Big Ideas Better Cities evenings on Health and Social Innovation through Big Data about “Combatting antibiotic resistance using surveillance”. See

A new NIH-funded collaboration with Dr. Larissa Williams, Bates College, USA that applies transcriptomic approaches to examination of the role Nfe2 in oxidative stress response during

http://traineeday2015.mcmasteriidr.ca

Wright, G.D. & A.G. McArthur. 2015. A bioinformatic platform for the characterization of antibiotic resistance in bacterial genomes and metagenomes. Presentation at the 2015 Interscience

I spent July travelling to two great meetings in the British Isles. First was the Galaxy Community Conference in Norwich, UK which provided a crash