More information: The IIDR & DBCAD Announce Funding for Collaborative Projects
Category: antibiotic resistance

New CARD Team Member!
The McArthur lab welcomes new Curator-Developer William Huynh to the Comprehensive Antibiotic Resistance Database staff. Looking forward to pushing CARD forward with William’s help!

The McArthur lab welcomes back summer students Rachel Tran & Arman Edalatmand plus first timers Marcel Jansen & Emily Panousis! These four will be covering

Welcome Ashraf Bazan, who has joined the lab and the McMaster Biochemistry & Biomedical Sciences graduate program! A familiar face in the IIDR as he started

Congratulations finishing thesis students!
COVID19 kept us from celebrating together, but we are very proud of the work you have accomplished! Rachel Tran | Biochem 4T15 – Exploring the
McArthur Lab & CARD during SARS-CoV-2
Quick update on the status of the McArthur Lab and the Comprehensive Antibiotic Resistance Database. While our home institution McMaster University is closed to on-site

Chen CY, Clark CG, Langner S, Boyd DA, Bharat A, McCorrister SJ, McArthur AG, Graham MR, Westmacott GR, Van Domselaar G Proteomics Clin Appl. 2019

CARD 2020: antibiotic resistome surveillance with the Comprehensive Antibiotic Resistance Database
Alcock BP, Raphenya AR, Lau TTY, Tsang KK, Bouchard M, Edalatmand A, Huynh W, Nguyen A-LV, Cheng AA, Liu S, Min SY, Miroshnichenko A, Tran

Guitor AK, Raphenya AR, Klunk J, Kuch M, Alcock B, Surette MG, McArthur AG, Poinar HN, Wright GD. Editor’s Pick! Antimicrobial Agents and Chemotherapy 2019 Oct

Waglechner N, McArthur AG, Wright GD. Nat Microbiol. 2019 Aug 12. [Epub ahead of print] Glycopeptide antibiotics are produced by Actinobacteria through biosynthetic gene clusters

David Braley Centre for Antibiotic Discovery gives researchers a fighting chance against antimicrobial resistance. A forward-looking McMaster donor is investing $7 million in a new

Empowering conventional antibiotics with new drug leads targeting the outer-membrane of Gram-negative pathogens. E. Brown (PI), G.D. Wright (co-I), B. Coombes (co-I), A.G. McArthur (co-I),
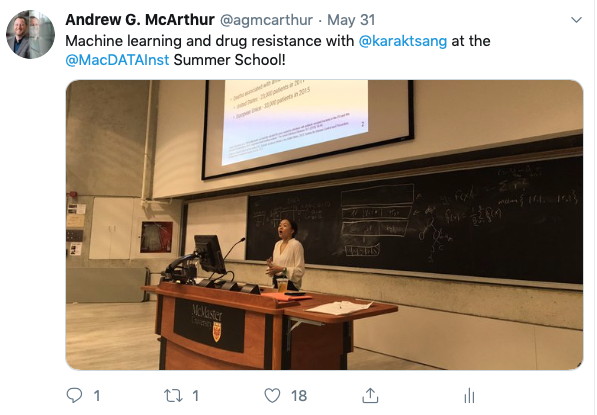
MacData Summer School 2019
Dr. McArthur and PhD student Kara Tsang taught together at the 2019 MacData Institute Summer School, with Dr. McArthur reviewing biocuration and bioinformatics for genomic

April 2019 big #card_release #RGI5! Resistance Gene Identifier Version 5: entirely new algorithms for metagenomics data, new options for genomes & assemblies. Extensively updated documentation,
The Comprehensive Antibiotic Resistance Database has been updated, http://card.mcmaster.ca CARD Curation: Expanded MCR, OXA & IMP beta-lactamase, and macrolide phosphotransferase (MPH) sequence curation. Updated nomenclature
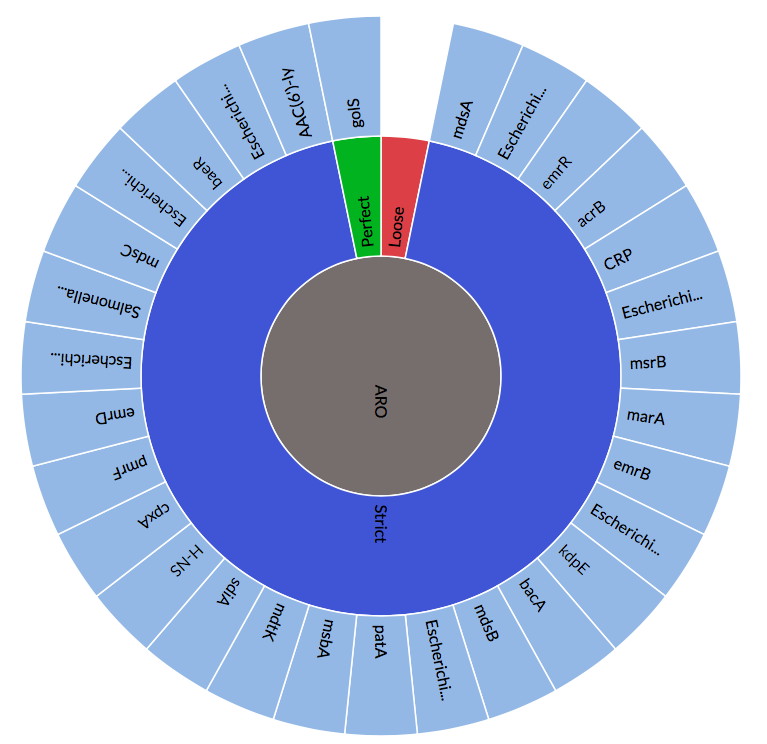
State of the CARD 2019
A week of lectures, demos, and training for the Comprehensive Antibiotic Resistance Database During McMaster Spring Mid-Term Recess (February 18-24), the McArthur lab is pleased

Antimicrobial Resistance: Emergence, Transmission, and Ecology (ARETE). R. Beiko (PI; Dalhousie University), F. Brinkman (co-PI, Simon Fraser University), A.G. McArthur (co-Applicant) + 4 additional co-Applicants.

Some invitations are more special than others. Dr. Peixoto da Cruz and I went to graduate school together in British Columbia (a long time ago!)
Maguire, F., B. Alcock, F.S. Brinkman, A.G. McArthur, & R.G. Beiko. 2018. AMRtime: Rapid Accurate Identification of Antimicrobial Resistance Determinants from Metagenomic Data. Oral presentation

