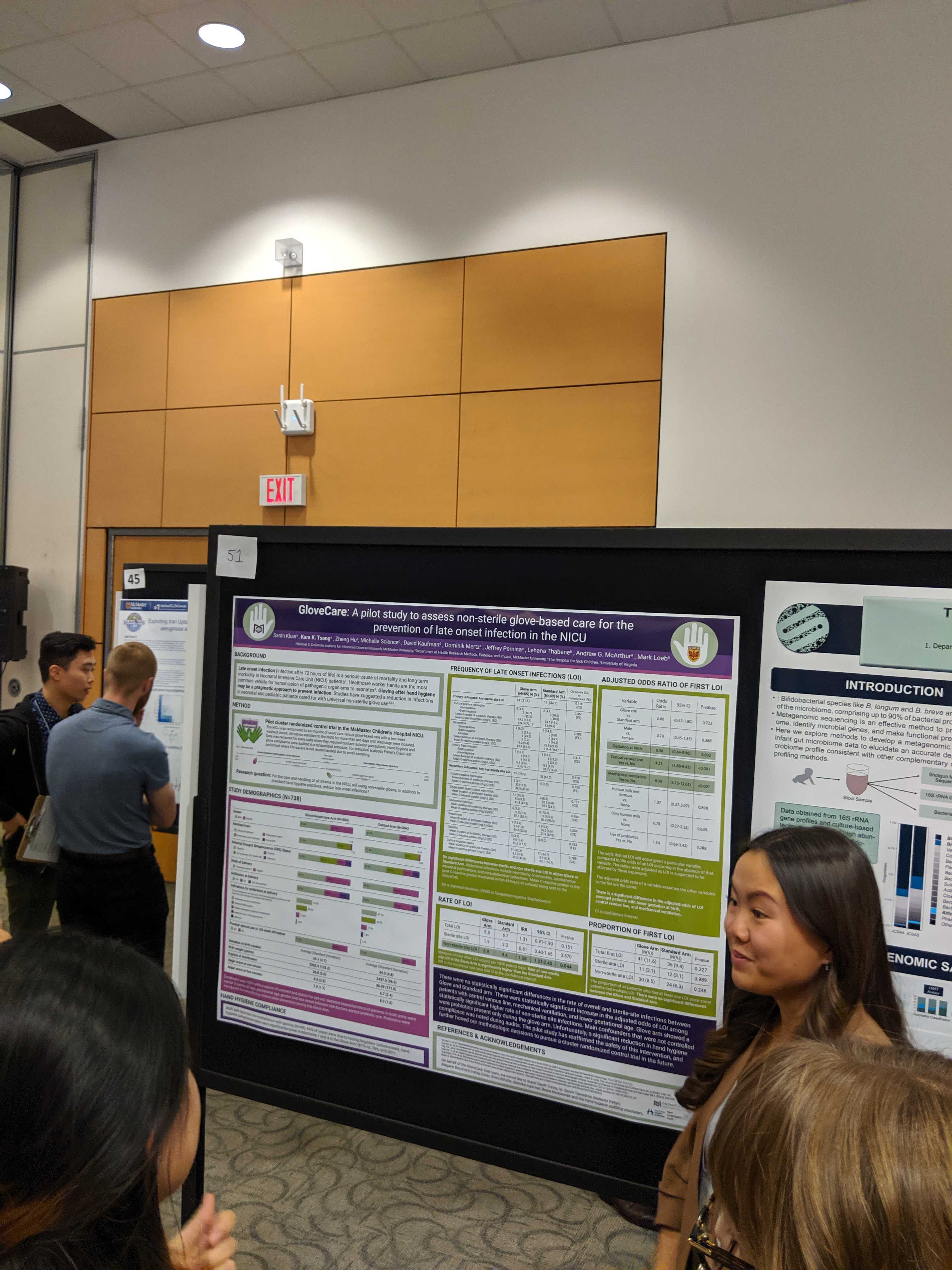
A great day of posters and presentations by the trainees of the Michael G. DeGroote Institute of Infectious Disease Research! Nilasha Mohan, Undergraduate student. Developing
Category: IIDR

This one-hour webinar, entitled “DNA sequencing in infectious disease surveillance, diagnosis, and management,” features talks from Andrew McArthur (Professor, Biochemistry and Biomedical Sciences) and Rubayet

IIDR Trainee Day 2023
COLIN BRUCE – Investigating Fibre Degradation in the Infant Gut Microbiota ; DIRK HACKENBERGER – Was World War 2 Foundational to the Antimicrobial Resistance Crisis?
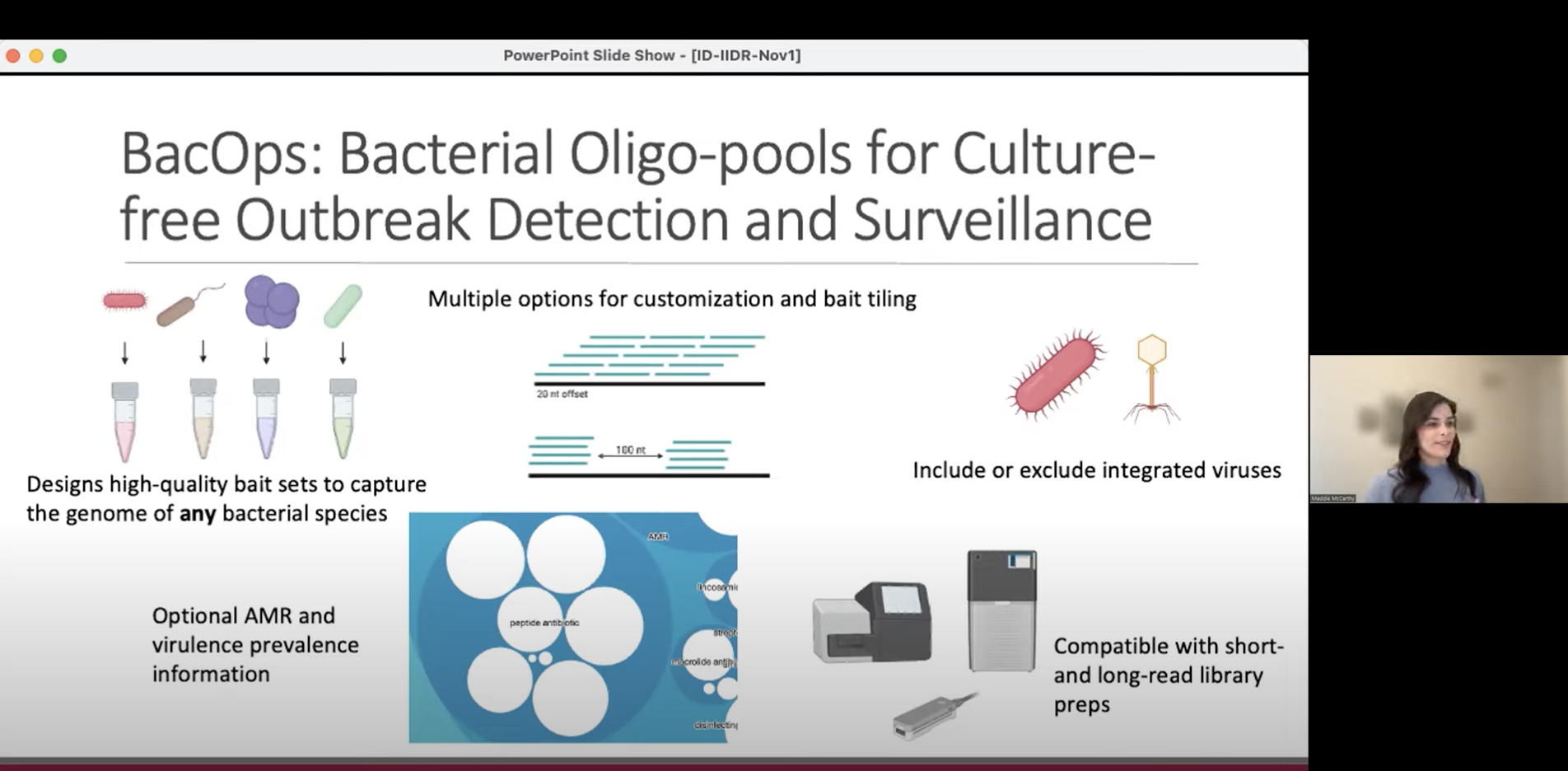
ID-IIDR Rounds – Clostridioides (Clostridium) difficile: new insights from the clinic and laboratory
This one-hour webinar features talks from Marek Smieja (Professor, Pathology & Molecular Medicine), Sheridan Baker (Postdoctoral Fellow, The McArthur Lab and Smieja Lab), and Maddie

IIDR Trainee Day 2022!
KARYN MUKIRI – Predicting the total resistome KEATON SMITH – Advancements in curation of the Comprehensive Antibiotic Resistance Database JALEES NASIR – Viral fishing expedition:

Congratulations Amogelang (Amos) Raphenya, winner of the 2022 Michael Kamin Hart Award (MSc). This award was created to honour the memory of Michael Kamin Hart,
Karyn Mukiri – Predicting the Total Resistome Mugdha Dave – Creation and standardization of automated bacterial pathogen genomic sequencing reporting through informatics

The McArthur lab welcomes Madeline McCarthy, a recent MSc graduate of VIDO / USask, as she join us for her PhD studies in the McMaster

Emily Panousis wins the Michael Kamin Hart Memorial Undergraduate Scholarship for her work on SARS-CoV-2 surveillance!
More information about IIDR Trainee Day and Michael Kamin Hart here.

Jalees Nasir awarded 2021 Fred & Helen Knight Enrichment Award!
Congratulations to PhD student Jalees Nasir, who has been awarded the prestigious 2021 Fred & Helen Knight Enrichment Award, which recognizes exceptional promise in early

Congratulations Emily Panousis on being awarded a 2021 IIDR Summer Student Fellowship! For nine years now, these awards have supported trainees working in IIDR and

It is with great pleasure the McArthur lab announces that Lead Developer for the Comprehensive Antibiotic Resistance Database (CARD), Amos Raphenya, is starting his M.Sc.

IIDR Trainee Day 2020! Forging the Future of Infectious Disease Research
Nasir, J.A et al. 2020. SIGNALing the Need for Surveillance: A Comparison of Whole Genome Sequencing Methods for SARS-CoV-2. Edalatmand, A. & A.G. McArthur. 2020.

McMaster is launching The Global Nexus for Pandemics and Biological Threats, to ensure Canada and the world are better able to manage the human and

New CARD Team Member!
The McArthur lab welcomes new Curator-Developer William Huynh to the Comprehensive Antibiotic Resistance Database staff. Looking forward to pushing CARD forward with William’s help!

Congratulations finishing thesis students!
COVID19 kept us from celebrating together, but we are very proud of the work you have accomplished! Rachel Tran | Biochem 4T15 – Exploring the
Edalatmand, A. & A.G. McArthur. 2019. Classifying and curating publications on antimicrobial resistance using machine learning. Poster presentation at the Michael G. DeGroote Institute for

The McArthur Lab is very proud to announce Amos Raphenya as 2019 IIDR Michael Kamin Kart Memorial Scholarship Recipient (Staff) and Rachel Tran as 2019
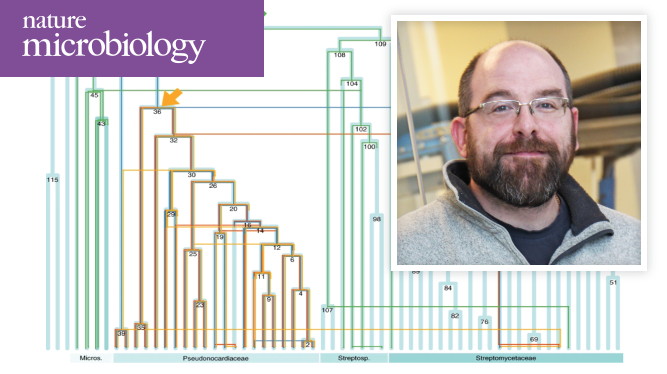
Waglechner N, McArthur AG, Wright GD. Nat Microbiol. 2019 Aug 12. [Epub ahead of print] Glycopeptide antibiotics are produced by Actinobacteria through biosynthetic gene clusters